At the heart of LabDAO is the idea that a community-operated open-source marketplace protocol for laboratory services could 1) accelerate progress and 2) generate a dynamic knowledge graph for biomedicine, where the receipts of past transactions among scientists serve as its building blocks.
To create the marketplace protocol and the subsequent knowledge graph, we need to work out a structured way to describe scientific services and the data they generate. We need standards for composable metadata.
Scientific data should be FAIR
Scientific data is ideally stored according to FAIR principles, developed by Mark Wilkinson. These include:
-
Findability
-
Accessibility
-
Interoperability
-
Reuse
Put differently, data is ideally stored at a location that is known to others, accessible to others, adheres to public standards, and thus invites reuse.
Web3 introduces new methods to share, control and distribute data according to FAIR principles. These methods include:
-
direct content addressing using IPFS
-
on-chain provenance tracking using NFTs
-
robust access control using tools like threshold encryption
With these methods, data can be pinned visibly and addressable for everyone. Ownership of data can be traced and access to encrypted information can be tied to token ownership. Web3 increases the findability and accessibility of data - the "F" and "A" of FAIR.
To make data more interoperable ("I") and reuseable ("R") its formatting needs to be standardized and it needs to be presented with sufficient metadata to provide relevant context. LabDAO is an online community of scientists and engineers that, among many things, define standards for data formats and metadata structure. Here we introduce a proposed simple standard for tokenized scientific data.
Web3 x Bio forms of metadata
Let's take a look at common metadata formats seen in both web3 and bio.
NFTs
A common case where metadata in web3 is important is for NFTs following the ERC721 standard. An object, often a digital artwork, is referenced within a metadata JSON together with additional properties. Often these properties give us more details about the artwork.
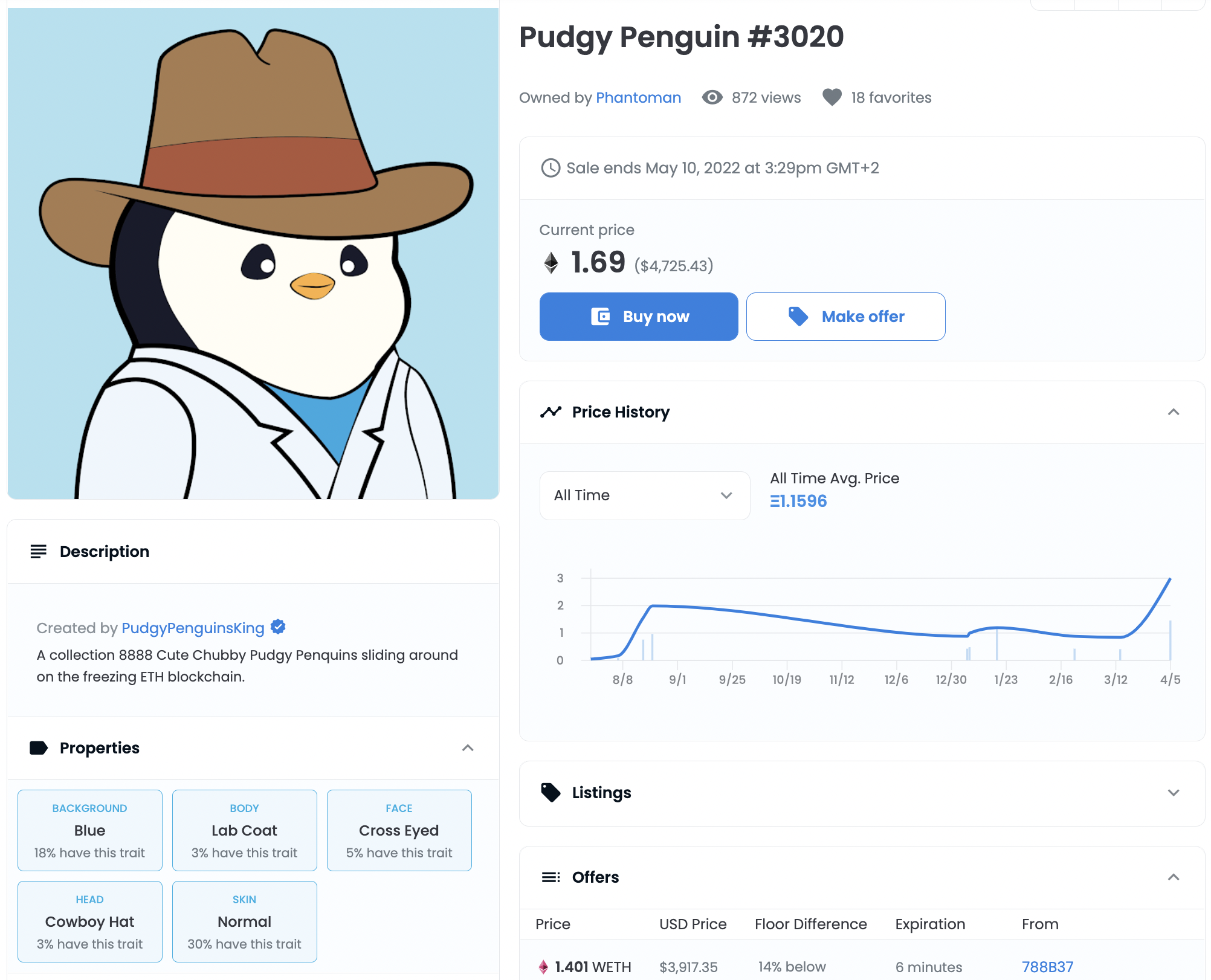
Below you can see the metadata JSON file for our example. The picture of the penguin itself is referenced under image using a universal resource identifier (URI).
{
"attributes": [
{
"trait_type": "Background",
"value": "Blue"
},
{
"trait_type": "Skin",
"value": "Normal"
},
{
"trait_type": "Body",
"value": "Lab Coat"
},
{
"trait_type": "Face",
"value": "Cross Eyed"
},
{
"trait_type": "Head",
"value": "Cowboy Hat"
}
],
"description": "A collection 8888 Cute Chubby Pudgy Penquins sliding around on the freezing ETH blockchain.",
"image": "https://ipfs.io/ipfs/QmNf1UsmdGaMbpatQ6toXSkzDpizaGmC9zfunCyoz1enD5/penguin/3020.png",
"name": "Pudgy Penguin #3020"
}
This existing NFT standard has also been adapted by other projects, such as the Molecule team to represent legal sublicenses for intellectual property. This form of NFT is called an IP-NFT.
Biocompute Objects
In Bio, specifically computational biology, an international IEEE standard for metadata exists biocompute objects. If this is absolutely new to you, you are in good company. A lot of biologists, especially in academia, do not know this standard exists.
To adhere to this standard (which rarely happens), scientists have to provide extensive information about multiple aspects:
-
who created the data (provenance domain)
-
a description of the data (usability domain)
-
a step-by-step computational pipeline of how the data was processed (description domain)
-
information about the environment in which the processing was run (execution domain)
-
links to input and output data (io domain)
-
parameters that were used in the computational pipeline (parameter domain)
Biocompute objects like this JSON can easily take up more than 700 lines. Biocompute objects lack any web3 primitives, such as decentralized identifiers and stable content addressing via IPFS for referenced data. Could there be a simplified structure for biomedical metadata that makes use of all the tools decentralized storage and ledgers give us?
introducing lab-NFTs
By building on the basic structure of the biocompute object and simplifying it with web3 primitives, we have come up with an internal standard for lab-NFT metadata. To reduce the complexity of any given metadata object and allow for reusability of components -a prerequisite for knowledge graphs- we factorize the metadata into 7 components:
-
general object information, including the standardized name of the requested service and a simple description
-
user information (user object)
-
parameters for the service (parameter object)
-
input data which includes a description and links to a file (input object)
-
provider information (provider object)
-
execution information which contains information about the runtime and the computational graph that was executed. The execution is often pointing to a community-maintained container image (execution object)
-
output data which includes a description and links to a file (output object)
Components 2-7 are all pinned as separate JSON objects on IPFS and referenced here. Their URI can be reused in other transactions, leading to new lab-NFTs, and a branched tree of references - our knowledge graph.
A very simple example of metadata can be seen below. In this example, a user is requesting the marketplace to generate the reverse complement of a set of DNA sequences.
{
"name": "reverse-complement", # a standardized name for the performed process
"description": "the reverse complement of a DNA sequence",
"image": "ipfs://QmZ9oReVUiNQSc9GaqqTEPUW3XHo6eprVSa9nqbGNotP8B",
"properties": {
"user": "0x64BC15E0A5A12dDbe321EEDD832d057775D11F56"
"parameters": # no parameters in this simple example
"input": "ipfs://QmaQv91AUmu4wDNiNc8exUrSrDnZ65YBEXuTrxSJSnJbcz",
"provider": # provider wallet address goes here
"execution": # object URI containing details about the execution environment goes here
"output": # object URI containing a referenced .fasta file
}
}
To initiate the transaction, the user is generating an incomplete lab-NFT metadata object, which we refer to as a job object. A job object includes the top-level descriptors, including the name of the service. In addition, the job object includes a reference to the 1) user, 2) parameters and 3) input data.
Once the transaction request is posted and funds have been deposited in the exchange escrow, providers in the network can claim the job. After completing the service, the provider adds references to 4) their identity, 5) information about the job execution and 6) output data.
Simply put, a user of the protocol is creating a half-baked NFT metadata object and is paying someone else (the provider) to complete the metadata and mint the token. The token is an entry in our knowledge graph. Scientists work with other scientists and create entries in a global knowledge graph of scientific inquiry.
The user and provider object represent the identity of the people involved in the transaction. The identity can be a referenced JSON object or a holonym.
A parameter object is defined by the LabDAO community and is application-specific. Certain simple applications, such as our reverse complement example, do not require parameters.
An example for parameters of a more complex job, an alphafold v2 run, is shown below:
{
"weights": "ipfs://bafybeihjjbkgeyhpfrgpnqpidkoha5uvnqfdoffsy4ipnrxbpyfxcipwh4",
"mode": "monomer",
"database": "full",
"max_template_date": "2022-01-01",
"is_prokaryote": false
}
Returning to our reverse complement example, the input object is following a simple structure and is referencing a fasta file with the DNA sequence of interest. The output object added by the provider (after completion of the job) follows the same structure.
{
"name": "example_sequence.fasta"
"type": "fasta"
"uri": "ipfs://QmfURWZakhnD1Rn3DqC38Eqps1zgZ17dxM4KgV1rmJDhww"
}
The execution object contains information about the environment in which a job was processed. In our case, it is referencing a container image used for the completion of the job.
{
"image" # URI of reverse complement application container
"runtime": "deta microservice, https://02wun6.deta.dev/"
"logs": # additional information
}
Endgame: encryption and composable NFTs
The lab-NFT concept allows us to exchange FAIR scientific data on the lab-exchange. Scientists can easily work together to perform research and share their results openly.
To accelerate not only progress in the basic sciences but also innovation around valuable intellectual property, we will introduce threshold encryption for lab-NFTs. All input and output data is symmetrically encrypted before it is pinned to IPFS. The owner of the lab-NFT can request access to the referenced files using tools like lit protocol.
Most scientific publications are based on data generated through multiple laboratory services. The same is true for most forms of intellectual property. To enable users to bundle individual lab-NFTs for simplified distribution, composable IP-NFTs will be developed based on existing implementations, such as aavegotchis which are inspired by the EIP998 proposal. Composability will allow authors to bundle their work, and distribute it more easily.
Get involved
If you are curious about what we are doing at LabDAO, join our discord and subscribe to our newsletter.
Thank you, Rik, Aakaash, Jesse, Clemens, and Lily for your thoughts on lab-NFT metadata.